| site | altitude | csize.mean | csize.sd | n |
|---|---|---|---|---|
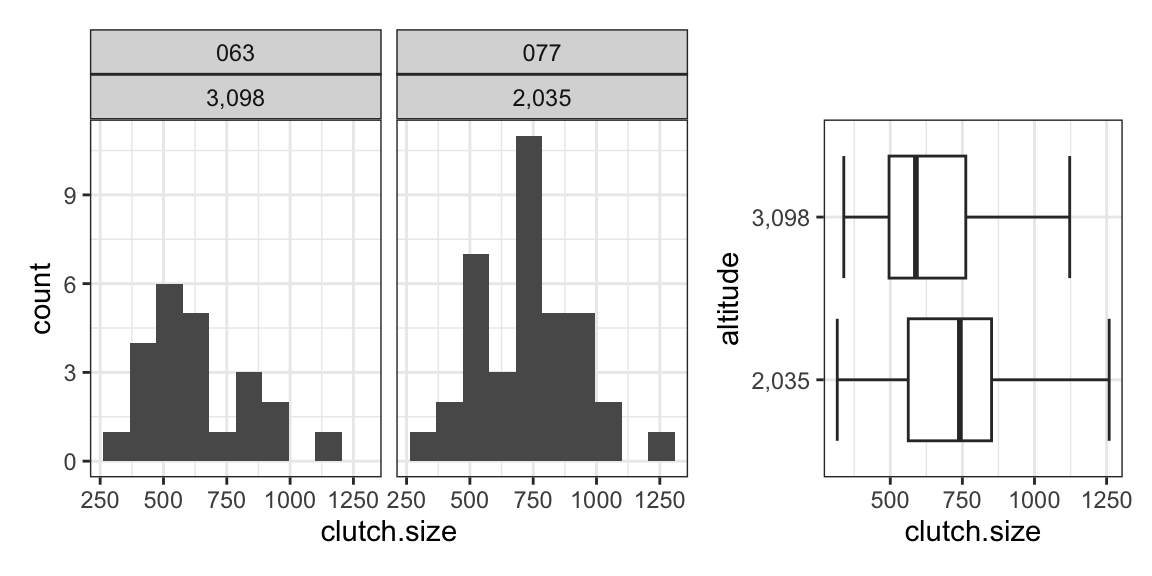
| 063 | 3,098 | 625.67 | 197.01 | 23 |
| 077 | 2,035 | 733.44 | 202.72 | 37 |
STAT218
February 4, 2025
The table and figures below show summary statistics and distributions of clutch sizes for two of the sites in the frog data from Test 1.

| site | altitude | csize.mean | csize.sd | n |
|---|---|---|---|---|
| 063 | 3,098 | 625.67 | 197.01 | 23 |
| 077 | 2,035 | 733.44 | 202.72 | 37 |
# perform t test
test.out <- frog |>
filter(site %in% c('077', '063')) |>
select(site, altitude, clutch.size) %>%
t.test(clutch.size ~ site, data = .)
# point estimate and std error
c(diff(test.out$estimate), test.out$stderr)mean in group 077
107.77512 52.89777 t
-2.037423 [1] -50.9182 266.4684At the 1% level, there is no evidence of a difference in mean clutch size between frog populations at the two sites.
The lizards data contains top running speeds in meters per second (m/s) from two species of lizard: western fence and sagebrush.
load('data/lizards.RData')
# side-by-side boxplots
boxplot(top.speed ~ species, data = lizards, horizontal = T)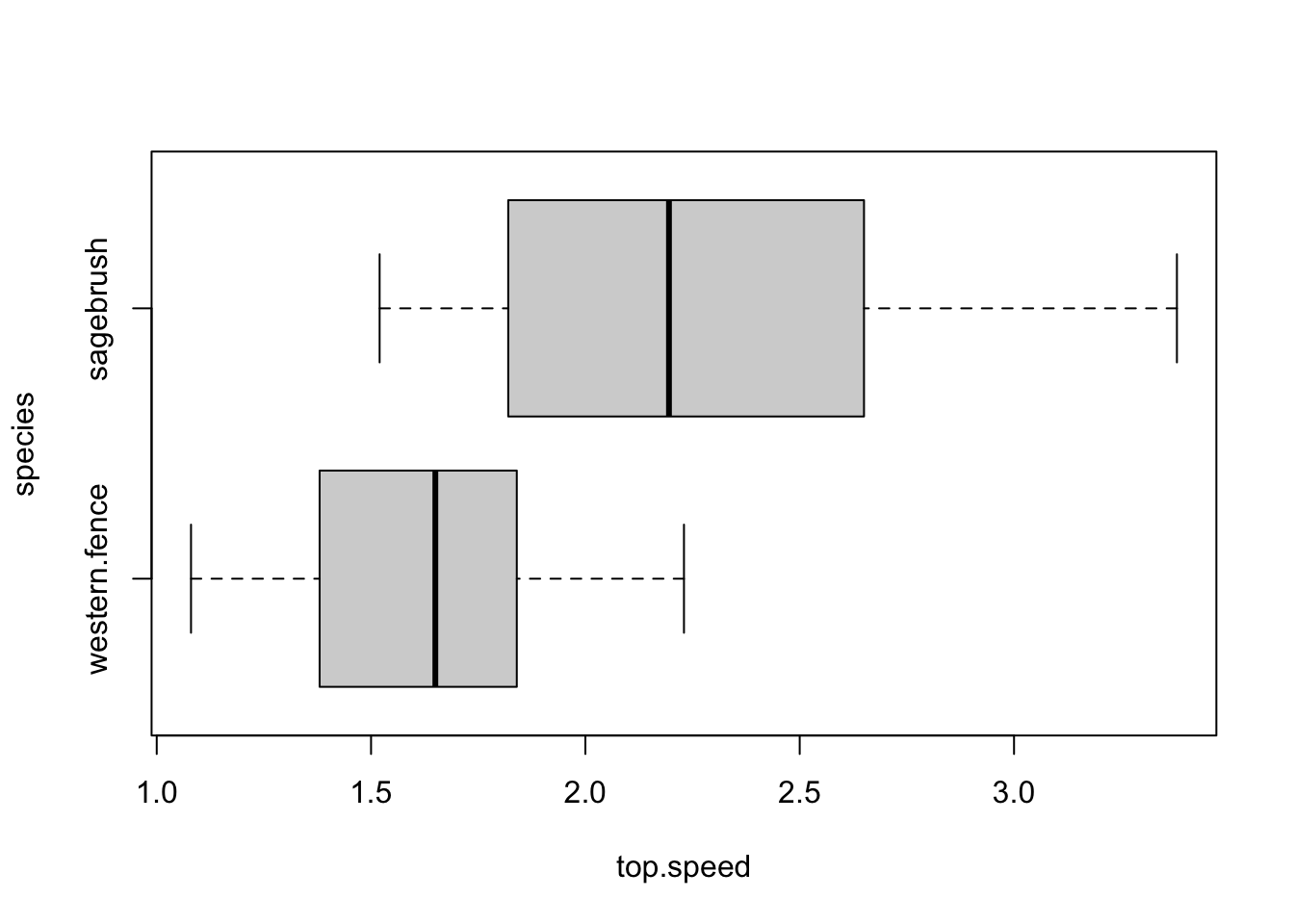
# test for a difference in mean top speed
tt.out <- t.test(top.speed ~ species, data = lizards)
tt.out
Welch Two Sample t-test
data: top.speed by species
t = -5.2217, df = 32.57, p-value = 9.939e-06
alternative hypothesis: true difference in means between group western.fence and group sagebrush is not equal to 0
95 percent confidence interval:
-0.9754526 -0.4282537
sample estimates:
mean in group western.fence mean in group sagebrush
1.612692 2.314545 diff.mean in group sagebrush se
0.7018531 0.1344115 [1] -0.9754526 -0.4282537
attr(,"conf.level")
[1] 0.95