library(tidyverse)
library(effectsize)
library(emmeans)Homework 6
In a study1, female mice were randomly assigned to four treatment groups to investigate whether restricting dietary intake increases life expectancy:
[NP] mice ate unlimited amount of nonpurified, standard diet
[N/N85] normal diet before weaning and normal diet after weaning (85 kcal/wk)
[N/R50] normal diet before weaning and reduced calorie diet after weaning (50 kcal/wk)
[N/R40] normal diet before weaning and reduced diet after weaning (40 Kcal/wk)
The
longevitydataset contains observations of lifetime in weeks for 237 mice from the study described above. In this problem you’ll test whether diet restriction has an effect on longevity.- Make side-by-side boxplots of the lifespan data in each treatment group. Assess whether the model assumptions for ANOVA seem plausible.
- Compute summary statistics by treatment group: sample means, standard deviations, sample sizes, and standard errors for the sample means.
- Test for an effect of diet restriction on mean lifespan. Interpret the result of the test in context following the narrative format from class.
- Estimate the effect size of diet restriction on mean lifespan; provide and interpret a (two-sided) 95% confidence interval.
# read in data and preview
load('data/longevity.RData')
head(longevity) lifetime diet
1 35.5 NP
2 35.4 NP
3 34.9 NP
4 34.8 NP
5 33.8 NP
6 33.5 NP# part a: side-by-side boxplots
boxplot(lifetime ~ diet, data = longevity)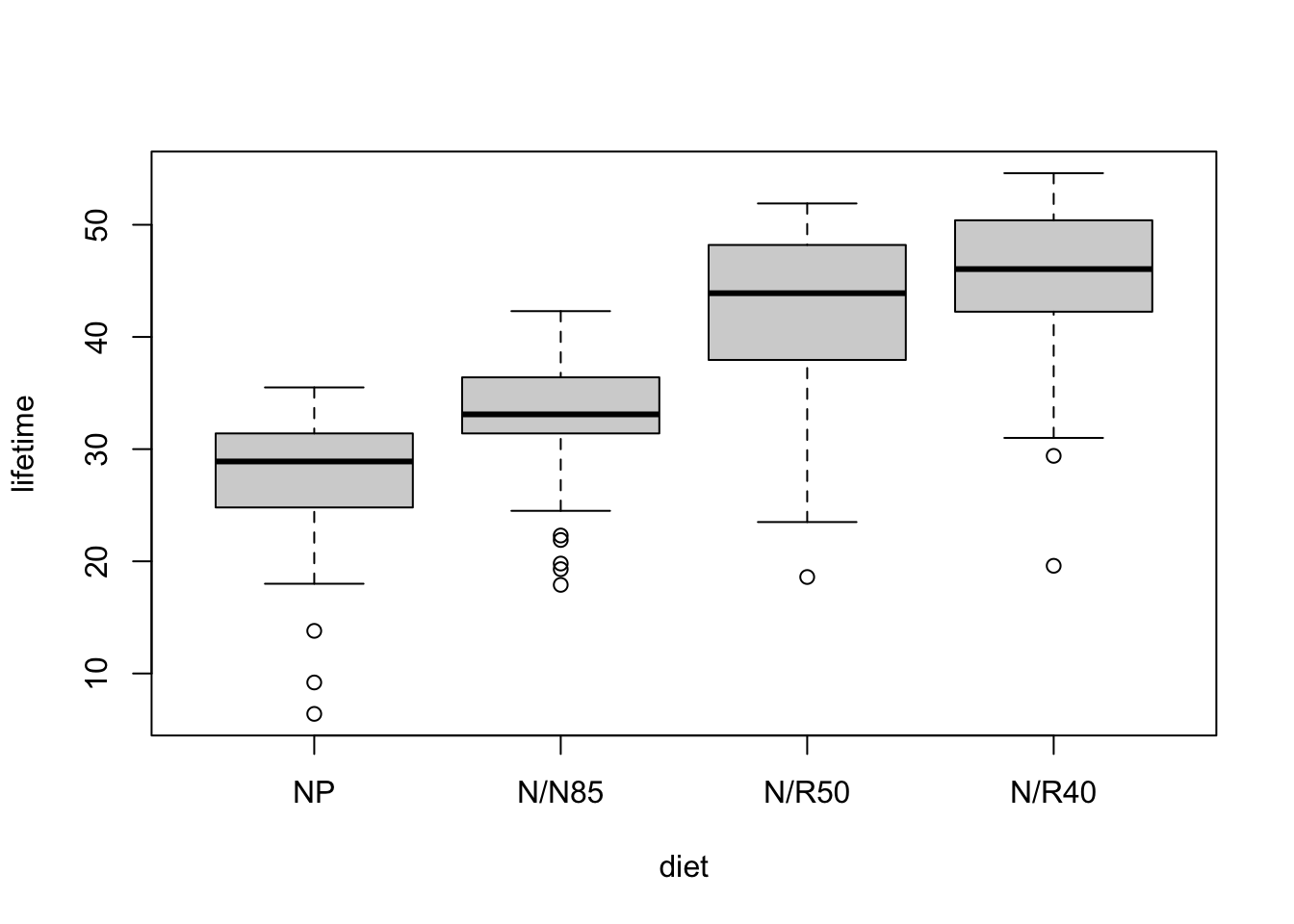
# part b: summary statistics by treatment group
longevity |>
group_by(diet) |>
summarize(mean = mean(lifetime),
sd = sd(lifetime),
n = length(lifetime),
se = sd/sqrt(n))# A tibble: 4 × 5
diet mean sd n se
<fct> <dbl> <dbl> <int> <dbl>
1 NP 27.4 6.13 49 0.876
2 N/N85 32.7 5.13 57 0.679
3 N/R50 42.3 7.77 71 0.922
4 N/R40 45.1 6.70 60 0.865# part c: omnibus test for effect of diet on mean lifespan
fit <- aov(lifetime ~ diet, data = longevity)
summary(fit) Df Sum Sq Mean Sq F value Pr(>F)
diet 3 11426 3809 87.41 <2e-16 ***
Residuals 233 10152 44
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# part d: estimate effect size
eta_squared(fit, partial = F, alternative = 'two.sided', ci = 0.95)# Effect Size for ANOVA (Type I)
Parameter | Eta2 | 95% CI
-------------------------------
diet | 0.53 | [0.44, 0.60]- The distributions show similar variability across groups, and individually data within groups satisfies
- The data provide evidence that caloric intake affects mean lifespan (F = 87.41 on 3 and 233 df, p < 0.0001).
- With 95% confidence, an estimated 44-60% of variation in lifespans is attributable to caloric intake.
Imagine you are designing a follow up study on dietary restriction.
- If there are the same number of levels of dietary restriction as in the original study, how many mice per group would you need to detect an effect size of
- If there are only two levels of dietary restriction, and you want to detect a difference in mean lifespan of 1 week or more 90% of the time using a 5% level test, how many mice do you need in your study? (Round the largest standard deviation from the original study to the next nearest whole number for a conservative estimate.)
- If there are six levels of dietary restriction, how many mice per group would you need to detect an effet of the same magnitude as estimated in the original study 85% of the time with a 5% level test?
- If there are the same number of levels of dietary restriction as in the original study, how many mice per group would you need to detect an effect size of
# part a: same number of levels, effect size 0.2
power.anova.test(groups = 4,
sig.level = 0.05,
power = 0.8,
between.var = 0.2,
within.var = 0.8)
Balanced one-way analysis of variance power calculation
groups = 4
n = 15.54913
between.var = 0.2
within.var = 0.8
sig.level = 0.05
power = 0.8
NOTE: n is number in each group# part b: two levels, difference of 1 week
power.t.test(delta = 7,
power = 0.9,
sd = 8,
sig.level = 0.05)
Two-sample t test power calculation
n = 28.44356
delta = 7
sd = 8
sig.level = 0.05
power = 0.9
alternative = two.sided
NOTE: n is number in *each* group# part c: six levels, effect size same as in original study
power.anova.test(groups = 6,
sig.level = 0.05,
power = 0.85,
between.var = 0.53,
within.var = 0.47)
Balanced one-way analysis of variance power calculation
groups = 6
n = 3.602306
between.var = 0.53
within.var = 0.47
sig.level = 0.05
power = 0.85
NOTE: n is number in each group- [Extra credit] Continuing to refer to the longevity study above, follow the example from lecture to compute interval estimates for log-contrasts and back-transform interval endpoints to obtain estimates for the percent change in median lifespan relative to the control group. Report the comparison between the normal (N/N85) diet and the unrestricted (NP) diet. (Note:
log(...)in R computes the natural logarithmexp(...)computes the exponential
load('data/longevity.RData')
# fit anova model to log lifetimes
fit.log <- aov(log(lifetime) ~ diet, data = longevity)
# estimate contrasts with control
emmeans(fit.log, ~ diet) |>
contrast('trt.vs.ctrl') |>
confint(level = 0.95, adjust = 'dunnett') contrast estimate SE df lower.CL upper.CL
(N/N85) - NP 0.200 0.0434 233 0.0972 0.303
(N/R50) - NP 0.452 0.0413 233 0.3538 0.550
(N/R40) - NP 0.524 0.0429 233 0.4217 0.625
Results are given on the log (not the response) scale.
Confidence level used: 0.95
Conf-level adjustment: dunnettx method for 3 estimates # back-transform point estimate for n85/np contrast
exp(0.200)[1] 1.221403# back-transform interval estimates
c(exp(0.097), exp(0.303))[1] 1.101860 1.353914With 95% confidence, median lifespan is an estimated 10.2% and 35.4% longer among mice on an 85kCal diet relative to an unrestricted calorie diet.
The
plantgrowthdataset includes measurements of dry weight of plants grown using one of two fertilizer treatments or no fertilizer (control); treatments were randomly allocated to plants.- Construct side-by-side boxplots of the data to assess ANOVA model assumptions.
- Fit an ANOVA model and test for a difference in mean dry weight among treatment groups at the 5% significance level. Report the results in context following conventional style.
- Estimate the effect size of fertilizer treatments on dry weight; provide a two-sided 95% confidence interval and interpret the interval in context.
- Test for significant differences in mean dry weight between each treatment compared with the control at the 5% level. Identify any significant differences.
- How do you explain the apparent discrepancy between the omnibus test and the post-hoc comparisons?
# load and inspect data
load('data/plantgrowth.RData')
head(plantgrowth) weight group
1 4.17 ctrl
2 5.58 ctrl
3 5.18 ctrl
4 6.11 ctrl
5 4.50 ctrl
6 4.61 ctrl# construct side-by-side boxplots
boxplot(weight ~ group, data = plantgrowth)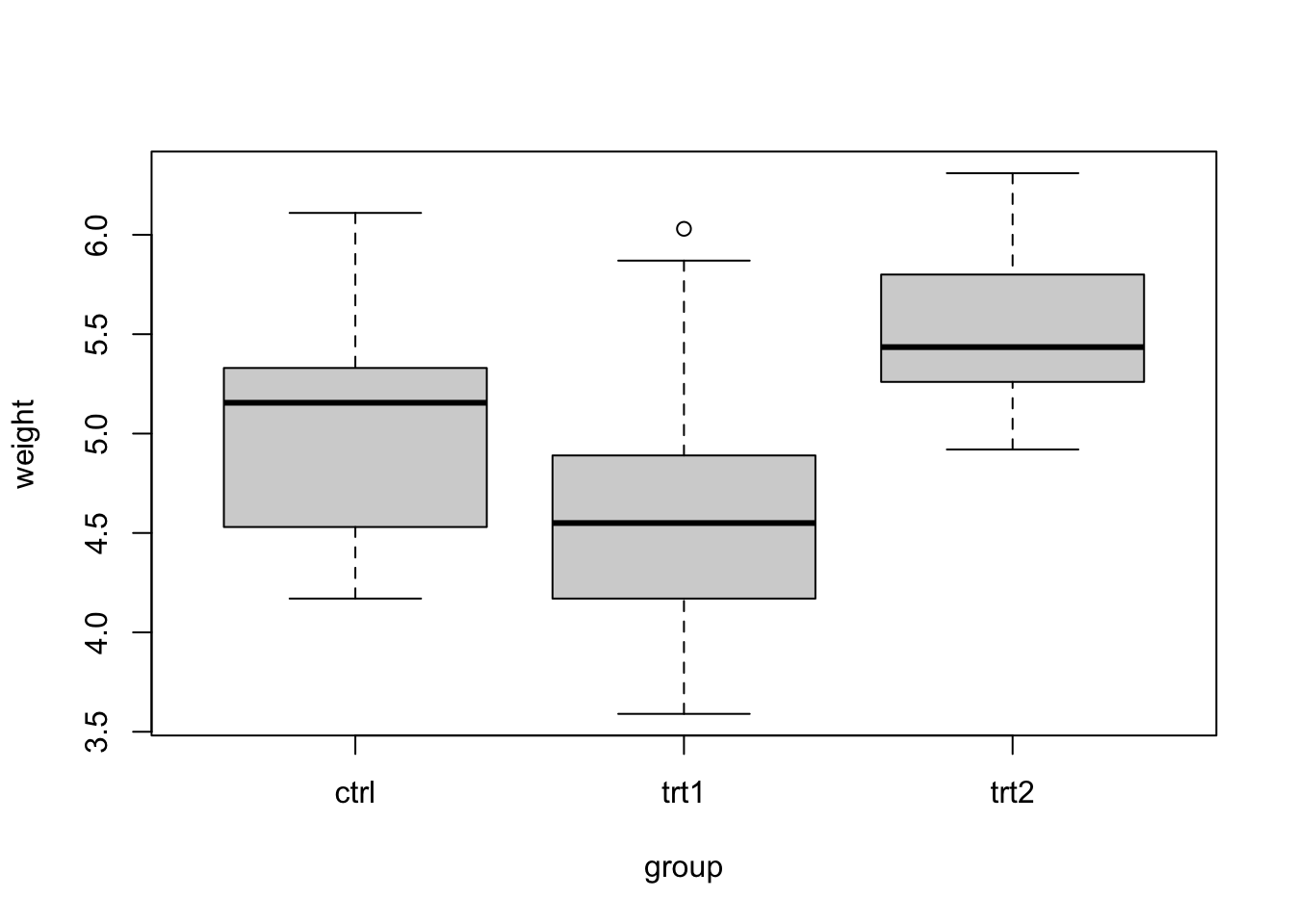
# fit anova model and perform omnibus test
fit.plant <- aov(weight ~ group, data = plantgrowth)
summary(fit.plant) Df Sum Sq Mean Sq F value Pr(>F)
group 2 3.766 1.8832 4.846 0.0159 *
Residuals 27 10.492 0.3886
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# estimate effect size
eta_squared(fit.plant, alternative = 'two.sided')# Effect Size for ANOVA
Parameter | Eta2 | 95% CI
-------------------------------
group | 0.26 | [0.01, 0.49]# test for contrasts with control
emmeans(fit.plant, ~ group) |>
contrast('trt.vs.ctrl') |>
test(adjust = 'dunnett') contrast estimate SE df t.ratio p.value
trt1 - ctrl -0.371 0.279 27 -1.331 0.3296
trt2 - ctrl 0.494 0.279 27 1.772 0.1582
P value adjustment: dunnettx method for 2 tests - The distributions show similar variability, and individually
- The data provide evidence that fertilizer treatment affects mean dry weight (F = 4.846 on 2 and 27 df, p = 0.0159).
- With 95% confidence, an estimated 1%-49% of variation in mean dry weight is attributable to fertilizer treatment.
- Neither treatment differs significantly from the control.
- The treatments differ significantly from each other, but not from the control.
Footnotes
Weindruch, R., Walford, R.L., Fligiel, S. and Guthrie D. (1986). The Retardation of Aging in Mice by Dietary Restriction: Longevity, Cancer, Immunity and Lifetime Energy Intake, Journal of Nutrition 116(4):641–54.↩︎