load('data/prevend.RData')
load('data/mammals.RData')Lab 11: Simple linear regression
The goal of this lab is to learn how to fit an SLR model, inspect the model summary, and generate confidence and prediction intervals. We’ll use the same datasets as last time.
Fitting an SLR model
To fit a linear model in R, use the lm(...) function:
# fit SLR model
fit.prevend <- lm(rfft ~ age, data = prevend)The syntax here is y ~ x, which you can read as “y depends on x”. The names that you provide in the formula must match variable names in the dataset identified by the data = ... argument.
To print the least squares estimates of the model coefficients:
# print coefficient estimates
coef(fit.prevend)(Intercept) age
134.098052 -1.190794 Each additional year of age is associated with a 1.19-point decrease in mean RFFT score.
Use lm(...) to fit a model to the mammal data relating log brain weight (y) to log body weight (x).
Store the model as fit.mammal. Print the coefficient estimates.
# fit SLR model
fit.mammal <- lm(log.brain ~ log.body, data = mammals)
# print coefficient estimates
coef(fit.mammal)(Intercept) log.body
2.1347887 0.7516859 Each 1-unit increment in log body weight is associated with a 0.75-unit increase in mean log brain weight.
If you wish to plot the fitted model over the data scatter, first make the plot and then add a line. (You have to run the whole cell for this to work.)
# visualize model
plot(prevend$age, prevend$rfft,
xlab = 'age', ylab = 'RFFT score')
abline(coef = coef(fit.prevend), col = 'blue')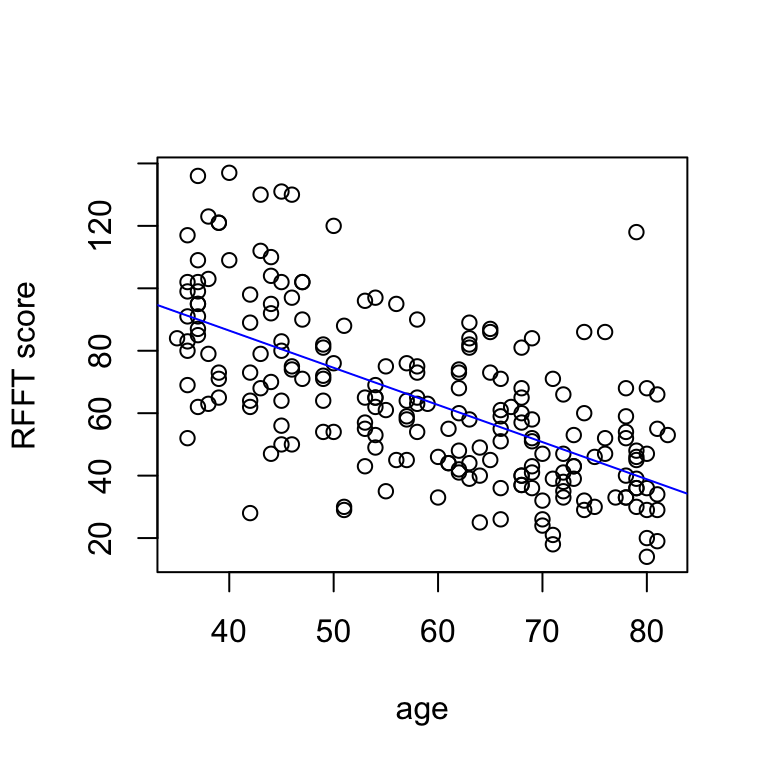
To obtain confidence intervals for the coefficients:
# confidence intervals for coefficients
confint(fit.prevend, level = 0.95) 2.5 % 97.5 %
(Intercept) 122.130647 146.0654574
age -1.389341 -0.9922471To see the model summary:
# model summary
summary(fit.prevend)
Call:
lm(formula = rfft ~ age, data = prevend)
Residuals:
Min 1Q Median 3Q Max
-56.085 -14.690 -2.937 12.744 77.975
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 134.0981 6.0701 22.09 <2e-16 ***
age -1.1908 0.1007 -11.82 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 20.52 on 206 degrees of freedom
Multiple R-squared: 0.4043, Adjusted R-squared: 0.4014
F-statistic: 139.8 on 1 and 206 DF, p-value: < 2.2e-16The model summary shows several key pieces of information:
- least squares estimates of coefficients and standard errors
- significance tests of whether each model parameter is zero
- estimated error variability
- proportion of variation explained by the model (
Take a moment to locate these.
Print the model summary for your model of brain and body weights, and locate and interpret the relevant pieces of information listed above.
# model summary
summary(fit.mammal)
Call:
lm(formula = log.brain ~ log.body, data = mammals)
Residuals:
Min 1Q Median 3Q Max
-1.71550 -0.49228 -0.06162 0.43597 1.94829
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.13479 0.09604 22.23 <2e-16 ***
log.body 0.75169 0.02846 26.41 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.6943 on 60 degrees of freedom
Multiple R-squared: 0.9208, Adjusted R-squared: 0.9195
F-statistic: 697.4 on 1 and 60 DF, p-value: < 2.2e-16The estimates are
The data provide evidence of an association between log body weight and log brain weight (
Log body weight explains an estimated 92.08% of log brain weight.
Predictions
To predict a new value for the response
# predicted rfft score for a 55 year old
predict(fit.prevend, newdata = data.frame(age = 55)) 1
68.60439 A typical domestic cat weighs 3.5-4.5kg. Estimate its brain weight using your SLR model.
Does the estimate seem close? (Google “cat brain weight”.)
Replace the ....
# predicted log brain weight
pred <- predict(fit.mammal,
newdata = data.frame(log.body = log(4)))
# predicted brain weight in grams
exp(pred) 1
23.97105 According to google, the average brain weight of a domestic cat is 25-30g. The point estimate is a bit low compared to this value.
To add either a confidence or prediction interval, append additional arguments for the interval type and the desired coverage level:
# confidence interval for mean rfft score
predict(fit.prevend, newdata = data.frame(age = 55),
interval = 'confidence', level = 0.95) fit lwr upr
1 68.60439 65.7101 71.49868# prediction interval for specific rfft score
predict(fit.prevend, newdata = data.frame(age = 55),
interval = 'prediction', level = 0.95) fit lwr upr
1 68.60439 28.04903 109.1598Fit gives the point estimate, and lwr and upr give the left and right interval endpoints, respectively.
Recall that these two intervals have different interpretations. For the confidence interval:
With 95% confidence, the mean RFFT score among 55-year olds is estimated to be between 65.71 and 71.50 points.
Whereas for the prediction interval:
With 95% confidence, the predicted RFFT score for a 55-year-old individual is estimated to be between 28.05 and 109.16 points.
Compute a confidence interval for your estimate from the last exercise. Does the interval capture the likely value?
Replace the ...
# predicted log brain weight
pred <- predict(fit.mammal,
newdata = data.frame(log.body = log(4)),
interval = 'confidence')
# predicted brain weight in grams
exp(pred) fit lwr upr
1 23.97105 20.09453 28.5954With 95% confidence, the brain weight of a typical 4kg domestic cat is estimated to be between 20.1 and 28.6 grams.
The confidence interval substantially overlaps with the range of 25-30 identified by internet search, so the estimate seems reasonable.
Now let’s have some fun with estimating brain weights.
Estimate your brain weight by converting your body weight to kg and using the SLR model. Construct a prediction interval.
Then google “human brain weight”. Does the prediction seem reasonable? If not, can you speculate about why not?
Replace the ...
# predicted log brain weight
pred <- predict(fit.mammal,
newdata = data.frame(log.body = log(68)),
interval = 'prediction')
# predicted brain weight in grams
exp(pred) fit lwr upr
1 201.65 49.25439 825.5651The prediction interval is far too low. Biologically, humans have a larger-than-typical brain relative to their body mass.
If you’re interested, have a look at the animals sorted by decreasing brain-to-body ratio.
# arrange by descending bb ratio
dplyr::arrange(mammals, desc(bb.ratio)) mammal body brain log.body log.brain bb.ratio
1 ground squirrel 0.101 4.00 -2.29263476 1.3862944 39.6039604
2 owl monkey 0.480 15.50 -0.73396918 2.7408400 32.2916667
3 lesser short-tailed shrew 0.005 0.14 -5.29831737 -1.9661129 28.0000000
4 rhesus monkey 6.800 179.00 1.91692261 5.1873858 26.3235294
5 galago 0.200 5.00 -1.60943791 1.6094379 25.0000000
6 little brown bat 0.010 0.25 -4.60517019 -1.3862944 25.0000000
7 mole rat 0.122 3.00 -2.10373423 1.0986123 24.5901639
8 tree shrew 0.104 2.50 -2.26336438 0.9162907 24.0384615
9 human 62.000 1320.00 4.12713439 7.1853870 21.2903226
10 mouse 0.023 0.40 -3.77226106 -0.9162907 17.3913043
11 baboon 10.550 179.50 2.35612586 5.1901752 17.0142180
12 star-nosed mole 0.060 1.00 -2.81341072 0.0000000 16.6666667
13 rock hyrax-a 0.750 12.30 -0.28768207 2.5095993 16.4000000
14 e. american mole 0.075 1.20 -2.59026717 0.1823216 16.0000000
15 chinchilla 0.425 6.40 -0.85566611 1.8562980 15.0588235
16 verbet 4.190 58.00 1.43270073 4.0604430 13.8424821
17 arctic fox 3.385 44.50 1.21935391 3.7954892 13.1462334
18 big brown bat 0.023 0.30 -3.77226106 -1.2039728 13.0434783
19 genet 1.410 17.50 0.34358970 2.8622009 12.4113475
20 red fox 4.235 50.40 1.44338333 3.9199912 11.9008264
21 patas monkey 10.000 115.00 2.30258509 4.7449321 11.5000000
22 raccoon 4.288 39.20 1.45582042 3.6686767 9.1417910
23 slow loris 1.400 12.50 0.33647224 2.5257286 8.9285714
24 chimpanzee 52.160 440.00 3.95431592 6.0867747 8.4355828
25 golden hamster 0.120 1.00 -2.12026354 0.0000000 8.3333333
26 echidna 3.000 25.00 1.09861229 3.2188758 8.3333333
27 cat 3.300 25.60 1.19392247 3.2425924 7.7575758
28 phalanger 1.620 11.40 0.48242615 2.4336134 7.0370370
29 musk shrew 0.048 0.33 -3.03655427 -1.1086626 6.8750000
30 rat 0.280 1.90 -1.27296568 0.6418539 6.7857143
31 roe deer 14.830 98.20 2.69665216 4.5870062 6.6217127
32 african giant pouched rat 1.000 6.60 0.00000000 1.8870696 6.6000000
33 arctic ground squirrel 0.920 5.70 -0.08338161 1.7404662 6.1956522
34 tree hyrax 2.000 12.30 0.69314718 2.5095993 6.1500000
35 mountain beaver 1.350 8.10 0.30010459 2.0918641 6.0000000
36 rock hyrax-b 3.600 21.00 1.28093385 3.0445224 5.8333333
37 guinea pig 1.040 5.50 0.03922071 1.7047481 5.2884615
38 rabbit 2.500 12.10 0.91629073 2.4932055 4.8400000
39 european hedgehog 0.785 3.50 -0.24207156 1.2527630 4.4585987
40 desert hedgehog 0.550 2.40 -0.59783700 0.8754687 4.3636364
41 yellow-bellied marmot 4.050 17.00 1.39871688 2.8332133 4.1975309
42 goat 27.660 115.00 3.31998733 4.7449321 4.1576283
43 grey seal 85.000 325.00 4.44265126 5.7838252 3.8235294
44 n.a. opossum 1.700 6.30 0.53062825 1.8405496 3.7058824
45 grey wolf 36.330 119.50 3.59264385 4.7833164 3.2892926
46 sheep 55.500 175.00 4.01638302 5.1647860 3.1531532
47 nine-banded armadillo 3.500 10.80 1.25276297 2.3795461 3.0857143
48 tenrec 0.900 2.60 -0.10536052 0.9555114 2.8888889
49 donkey 187.100 419.00 5.23164323 6.0378709 2.2394441
50 gorilla 207.000 406.00 5.33271879 6.0063532 1.9613527
51 okapi 250.000 490.00 5.52146092 6.1944054 1.9600000
52 asian elephant 2547.000 4603.00 7.84267147 8.4344635 1.8072242
53 kangaroo 35.000 56.00 3.55534806 4.0253517 1.6000000
54 jaguar 100.000 157.00 4.60517019 5.0562458 1.5700000
55 giant armadillo 60.000 81.00 4.09434456 4.3944492 1.3500000
56 giraffe 529.000 680.00 6.27098843 6.5220928 1.2854442
57 horse 521.000 655.00 6.25575004 6.4846352 1.2571977
58 water opossum 3.500 3.90 1.25276297 1.3609766 1.1142857
59 brazilian tapir 160.000 169.00 5.07517382 5.1298987 1.0562500
60 pig 192.000 180.00 5.25749537 5.1929569 0.9375000
61 cow 465.000 423.00 6.14203741 6.0473722 0.9096774
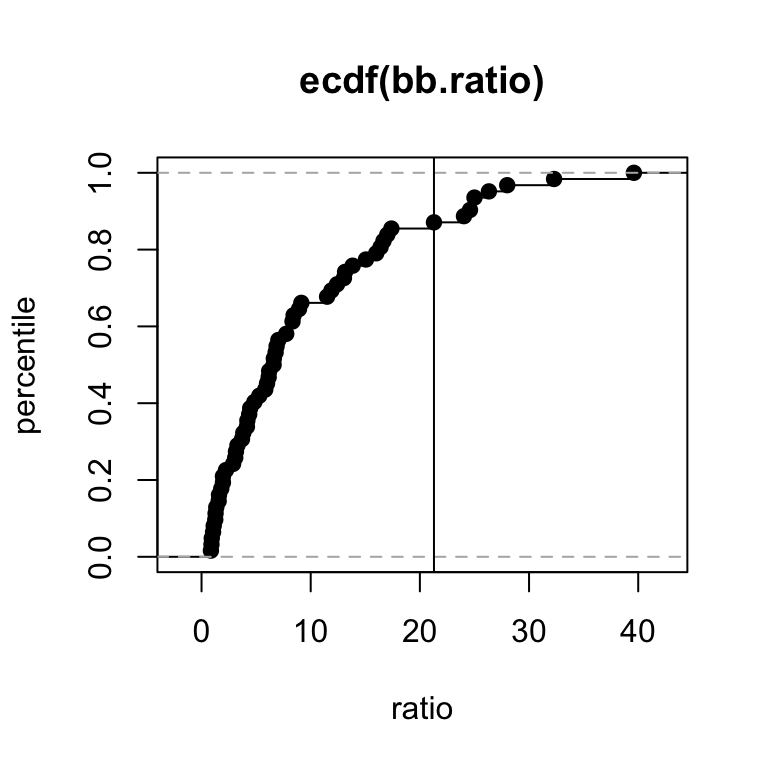
62 african elephant 6654.000 5712.00 8.80297346 8.6503245 0.8584310The model might not predict as well at the extremes of the brain-to-body ratio. In particular, note below that humans are about at the 85th percentile.
# humans are at about the 85th percentile of brain-to-body ratios
bb.ratio <- mammals$bb.ratio
ecdf(bb.ratio) |> plot(xlab = 'ratio', ylab = 'percentile')
abline(v = bb.ratio[32])